More Information
Submitted: December 22, 2021 | Approved: January 06, 2022 | Published: January 07, 2022
How to cite this article: Afrifah KA, Alexander Badu-Boateng A, Antwi-Akomeah S, Motey EE, Boampong E, et al. Genetic identification of three exhumed human remains at a hospital in Ghana: a forensic case report. J Forensic Sci Res. 2022; 6: 006-011.
DOI: 10.29328/journal.jfsr.1001030
Copyright License: © 2022 Afrifah KA, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Global filer; Short tandem repeats; Humerus; Kinship analysis; Forensics
Abbreviations: DNA: Deoxyribonuleic Acid; PCR: Polymerase Chain Reaction; STR: Short Tandem Repeat; Qpcr: Quantitative Polymerase Chain Reaction
Genetic identification of three exhumed human remains at a hospital in Ghana: a forensic case report
Kofi Adjapong Afrifah1,2*, Alexander Badu-Boateng1, Samuel Antwi-Akomeah1, Eva Emefa Motey1, Emmanuel Boampong1, David Agyemang Adjem1, Osei Owusu-Afriyie3 and Augustine Donkor4
1Forensic Science Laboratory, CID Headquarters, Ghana Police Service, Accra, Ghana
2Department of Biochemistry and Biotechnology, Kwame Nkrumah University of Science and Technology (KNUST), Kumasi, Ghana
3Department of Pathology, Ghana Police Hospital, Accra, Ghana
4Department of Chemistry, College of Basic and Applied Sciences, University of Ghana, Legon, Ghana
*Address for Correspondence: Dr. Kofi Adjapong Afrifah, Forensic Science Laboratory, CID Headquarters, Ghana Police Service, Accra, Ghana, Email: [email protected]
DNA identification is very important in cases of high decomposition of dead bodies, in which the bodies cannot be identified by physical means.
To compare the results of DNA typing, it is necessary to have related subjects with which to perform comparative analyses. Such tests are normally performed by comparing DNA profiles from people known to be immediate family members of the presumptive victim, such as parents or children because they share half of their genetic material with the unidentified.
We report on how DNA analysis was used to solve a case of mixed-up bodies at a local mortuary in Ghana, West Africa. Two families and three buried human remains were in contention in this case. The first body (E9) was buried three months before exhumation. The second body (E11) was buried two and a half months before exhumation whiles the third body (E10) was buried a month before exhumation. Exhibit E5 was taken from an alleged child of the deceased, E11. Toenails of the exhumed bodies were sampled by a pathologist and used for DNA extractions using the QIAamp DNA Investigator Kit.
Profiles from relatives were generated for comparison purposes. All samples gave a quality amount of genomic DNA after quantification. DNA was amplified with a GlobalFiler PCR amplification kit. Profiles from relatives were generated for comparison purposes.
The human remains (exhibit E11) cannot be excluded as the biological father of the child (exhibit E5) because they share common alleles at all 23 genetic loci. The applicable combined paternity index was 17218125604.492 assuming a prior probability of 0.5. The probability of paternity is 99.99999999%. Based on this relationship testing, one of the bodies was successfully identified and handed over to the family for re-burial.
DNA identification is very important when a body has reached a point of decomposition where it cannot be identified by physical means [1,2].
To compare the results of DNA typing, it is necessary to have related subjects with which to perform comparative analyses. Such tests are normally performed by comparison with the DNA profiles from people known to be immediate family members of the presumptive victim, such as parents or children because they share half of their genetic material with the unidentified body [2,3].
Bones and nails are important in identifying decomposed bodies compared to soft tissues. Although buccal swabs and saliva samples have been demonstrated to be stable at room temperature for over one and five years respectively, DNA from bones and nails DNA has been shown to be successful in SNP detection assays even when clippings are stored at room temperature for over 20 years [4]. This is because, during nail growth, cells differentiate into the nail plate and are filled with keratin. In this keratinization process, the cells undergo programmed cell death, which results in considerable DNA fragmentation. Once the nail root cells are keratinized (and the DNA fragmented) during nail growth, the keratin tissue probably protects the DNA from further damage because keratinous tissue makes the DNA less accessible to oxidants and does not contain water. Water in samples leads to DNA damage through hydrolytic deamination of cytosine [5].
The most widely used genetic markers for forensic DNA typing in most crime laboratories are autosomal short tandem repeat (STR) loci [6]. Commercially available STR kits, such as the SGM plus PCR Amplification Kit, AmpFℓSTR Identifiler PCR amplification kit, AmpFℓSTR Global Filer kit (Applied Biosystems, Foster City, California) or the PowerPlex 16 and PowerPlex 24 systems (Promega, Madison, Wisconsin) make use of a set of 10–24 STR loci to provide a high level of diversity and resolution for identity testing [7,8]. These kits, and STR loci, have been used widely for the identification of human remains as well as in relationship testing, such as relationship testing and family reconstructions.
We report on how one set of three different exhumed human remains stored at a local mortuary were genetically identified after a third family refused to claim a corpse handed to them by the mortuary authorities, insisting that the corpse was not that of their relative. Investigations by the mortuary authorities and the police led to the exhumation of three human remains at a local cemetery and kept at the mortuary for identification. Crime scene investigators sampled bones for forensic DNA Analysis since there was no soft tissue. Close relatives of the missing female (alleged son, and alleged mother) were invited for buccal swab sampling for comparative DNA analysis to determine the identity of the human remains.
Samples
- E9: Humerus bone samples of humans remain 1
- E10: Humerus bone samples of humans remain 2
- E11: Humerus bone samples of humans remain 3
Family A
- E1: Buccal swab of an alleged child of deceased
Family B
- E5: Buccal swab of an alleged child of deceased
- E3: Buccal swab of the alleged father of deceased
DNA extractions
Genomic DNA was extracted from 100.0mg of powdered bone samples from the deceased as well as buccal swab samples from alleged relatives of exhumed human remains using the QIAamp DNA Investigator Kit (Qiagen, Hilden, Germany) following the Manufacturer’s instructions, for genetic testing. Extracted samples were stored at -20 ℃ prior to further analysis.
DNA quantification
DNA extracted from the samples were quantified with the 7500 qPCR equipment using the QuantifilerTM Trio DNA amplification kit (Applied Biosystems, Foster City, CA) following the manufacturer’s protocol.
STR amplification and capillary electrophoresis
The extracted DNA from the samples were then amplified with the 9700 PCR machine using AmpFLSTRTM GlobalFilerTM PCR Amplification Kit (Applied Biosystems, Foster City, CA) following the manufacturer’s protocol. The amplified STR targets were electrophoresed in the 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA) and the generated STR profiles were analyzed using GeneMapper IDX version 1.5 software (Applied Biosystems, Foster City, CA), following the manufacturer’s protocol.
Statistical calculations
The paternity indexes were computed using allelic frequencies of the Black African race.
The letters p, q, and r in the “mother, child and Alleged Father” columns below represent allele frequencies in the co-dominant system [14].
| Child | Mother | Father | Likelihood Ratio |
| q | pq | q | 1/q |
| pq | p or pr | q | 1/q |
| q | q | q | 1/q |
| pq | p or pr | qr or pq | 1/2q |
| pq | pq | pq | 1/(p+q) |
| pq | pq | q | 1/(p+q) |
| pq | pq | qr | 1/(2p+2q) |
Where:
p = allele in child and in mother
q = allele in child and in father
r = allele not found in the child.
Probability of Paternity = CPI/CPI + 1
Where CPI = Combined Paternity Index
A prior probability of 0.5 was assumed for all calculations.
Allele Frequencies for computation of Likelihood Ratios and Combined Paternity Indexes were taken from published population data [13].
The obtained results are shown in Tables 1, 2, 3, 4 and Figures 1 and 2 respectively.
| Table 1: DNA Concentrations and IPC CT values from Real-time Quantification. | ||
| Sample Name | Concentration (ng/µl) | IPC CT Value |
| E9 (Humerus bone samples of humans remain 1) | 0.486 | 27.268 |
| E10 (Humerus bone samples of humans remain 2) | 0.294 | 27.412 |
| E11 (Humerus bone samples of humans remain 3) | 0.621 | 27.343 |
| E1 (Buccal swab of an alleged child of deceased) | 12.872 | 27.406 |
| E3 (Buccal swab of the alleged father of deceased) | 14.423 | 27.102 |
| E5 (Buccal swab of an alleged child of deceased) | 11.845 | 27.27 |
| Table 2: Typing results of the analyzed samples. | ||||
| STR locus | Human remains E9 (Humerus) |
E5 (alleged child of E9 ) | E1 (alleged child of E9) | E3 (alleged father of E9) |
| D8S1179 | 13,14 | 12,13 | 14,16 | 14 |
| D21S11 | 32,32.2 | 28,36 | 28,33.2 | 27,28 |
| D7S820 | 8,10 | 9,10 | 9,10 | 9,10 |
| CSF1PO | 10,12 | 10,11 | 7,12 | 10 |
| D3S1358 | 14,16 | 15,16 | 14,15 | 15,16 |
| TH01 | 6,7 | 7,8 | 7,8 | 6,7 |
| D13S317 | 11,13 | 11,12 | 12,13 | 12 |
| D16S539 | 11,13 | 11,12 | 11,14 | 12,14 |
| D2S1338 | 21 | 23,24 | 21,25 | 17,22 |
| D19S433 | 13,15 | 12,13 | 14 | 11,13 |
| vWA | 15 | 16,20 | 16,19 | 17 |
| TPOX | 9,11 | 10,11 | 10,11 | 7,9 |
| D18S51 | 15,19 | 17,18 | 18,19 | 17,19 |
| D5S818 | 13 | 11,13 | 12,13 | 11,13 |
| FGA | 22,23 | 21,24 | 24 | 22,25 |
| Y INDEL | 2 | - | 2 | 2 |
| DYS391 | 10 | - | 10 | 10 |
| D2S441 | 12,14 | 11,14 | 11 | 11,14 |
| D22S1045 | 17,18 | 10,16 | 11,15 | 14,20 |
| SE33 | 20,26.2 | 15,28.2 | 20,26.2 | 16,29.2 |
| D10S1248 | 13,15 | 13 | 13,16 | 13,15 |
| D1S1656 | 12,15 | 14 | 11,16 | 14 |
| D12S391 | 19,20 | 17,20 | 17,23 | 19,20 |
| AMEL | XY | XX | XY | XY |
| Table 3: Typing results of the analyzed samples. | ||||
| STR locus | Human remain E10 | E3(alleged father of E10) | E5(alleged child of E10) | E1 (alleged child of E10) |
| D8S1179 | 12,15 | 14 | 12,13 | 14,16 |
| D21S11 | 30,31 | 27,28 | 28,36 | 28,33.2 |
| D7S820 | 8,10 | 9,10 | 9,10 | 9,10 |
| CSF1PO | 9,12 | 10 | 10,11 | 7,12 |
| D3S1358 | 16,17 | 15,16 | 15,16 | 14,15 |
| TH01 | 8,10 | 6,7 | 7,8 | 7,8 |
| D13S317 | 12,13 | 12 | 11,12 | 12,13 |
| D16S539 | 11 | 12,14 | 11,12 | 11,14 |
| D2S1338 | 22,24 | 17,22 | 23,24 | 21,25 |
| D19S433 | 16 | 11,13 | 12,13 | 14 |
| vWA | 15,16 | 17 | 16,20 | 16,19 |
| TPOX | 10 | 7,9 | 10,11 | 10,11 |
| D18S51 | 15,16 | 17,19 | 17,18 | 18,19 |
| D5S818 | 11 | 11,13 | 11,13 | 12,13 |
| FGA | 18.2,27 | 22,25 | 21,24 | 24 |
| Y INDEL | 2 | 2 | - | 2 |
| DYS391 | 11 | 10 | - | 10 |
| D2S441 | 11,12.3 | 11,14 | 11,14 | 11 |
| D22S1045 | 17 | 14,20 | 10,16 | 11,15 |
| SE33 | 18,19 | 16,29.2 | 15,28.2 | 20,26.2 |
| D10S1248 | 14,15 | 13,15 | 13 | 13,16 |
| D1S1656 | 14,16 | 14 | 14 | 11,16 |
| D12S391 | 15,18 | 19,20 | 17,20 | 17,23 |
| AMEL | XY | XY | XX | XY |
| Table 4: Typing results of the analyzed samples. | |||||
| STR locus | Human remain E11 | E3(alleged father of E11) | E5 (alleged child of E11) | E1 (alleged child of E11) | Paternity Index |
| D8S1179 | 12,14 | 14 | 12,13 | 14,16 | 4.098 |
| D21S11 | 28 | 27,28 | 28,36 | 28,33.2 | 4.132 |
| D7S820 | 10 | 9,10 | 9,10 | 9,10 | 2.105 |
| CSF1PO | 11 | 10 | 10,11 | 7,12 | 4.505 |
| D3S1358 | 16 | 15,16 | 15,16 | 14,15 | 1.592 |
| TH01 | 7 | 6,7 | 7,8 | 7,8 | 1.623 |
| D13S317 | 12 | 12 | 11,12 | 12,13 | 1.376 |
| D16S539 | 11 | 12,14 | 11,12 | 11,14 | 2.079 |
| D2S1338 | 19,23 | 17,22 | 23,24 | 21,25 | 9.346 |
| D19S433 | 12,14 | 11,13 | 12,13 | 14 | 5.000 |
| Vwa | 16 | 17 | 16,20 | 16,19 | 3.704 |
| TPOX | 8,10 | 7,9 | 10,11 | 10,11 | 5.495 |
| D18S51 | 12,17 | 17,19 | 17,18 | 18,19 | 2.841 |
| D5S818 | 10,13 | 11,13 | 11,13 | 12,13 | 2.101 |
| FGA | 23,24 | 22,25 | 21,24 | 24 | 2.924 |
| Y INDEL | 2 | 2 | - | 2 | - |
| DYS391 | 10 | 10 | - | 10 | - |
| D2S441 | 12.3,14 | 11,14 | 11,14 | 11 | 2.128 |
| D22S1045 | 16 | 14,20 | 10,16 | 11,15 | 5.376 |
| SE33 | 11.2,28.2 | 16,29.2 | 15,28.2 | 20,26.2 | 5.208 |
| D10S1248 | 8,13 | 13,15 | 13 | 13,16 | 2.155 |
| D1S1656 | 13,14 | 14 | 14 | 11,16 | 2.137 |
| D12S391 | 17,20 | 19,20 | 17,20 | 17,23 | 3.185 |
| AMEL | XY | XY | XX | XY | |
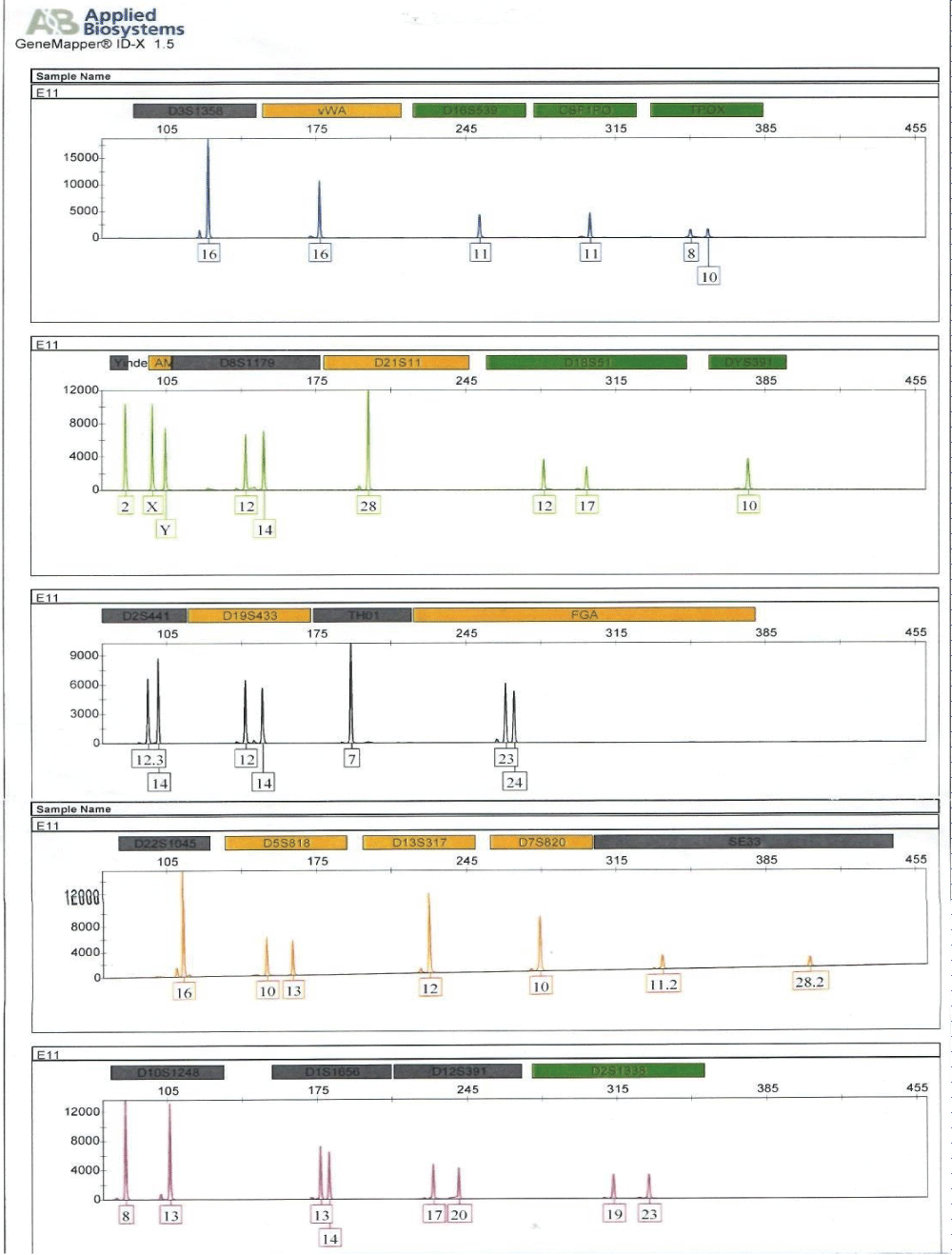
Figure 1: Electropherogram generated from short tandem repeat profiling of human remains, E11 (humerus bones).
Figure 2: Shows Electropherogram generated from short tandem repeat profiling of sample of an alleged child, E5.
DNA fingerprinting by STR loci analysis has become the ultimate method for the identification of human remains from mass disasters, armed conflicts and criminal investigations [9]. Of late, the application of improved or completely new laboratory protocols has greatly increased the success rate of nuclear DNA profiling of degraded human skeletal remains and affords forensic scientists and investigators the opportunity to accurately identify missing persons to bring closure to such cases for many families [10].
DNA profiling is very helpful in identifying human remains and in criminal investigations. DNA molecule is very unique to an individual and remains highly conserved in the person’s lifetime. Each individual’s genetic fingerprint is composed of equal parts of his or her parents’ DNA and can be analyzed to produce DNA profiles comparable to other DNA profiles of close relatives. DNA is a resilient molecule, degrades slowly in hard tissues, such as bones and teeth and can be recovered and analyzed from small crime scene biological samples, such as bloodstains, saliva, or single hair with roots [11].
The use of human remains sampled from the crime scene, and various taphonomic conditions usually result in small amounts of DNA being extracted for forensic analysis.
The DNA section of the Forensic Science Laboratory at the Criminal Investigation Department of the Ghana Police Service sampled human remains (humerus) from three human remains at a local hospital mortuary in the Western Region of Ghana. Buccal swabs of alleged relatives of the deceased whose remains were exhumed were also taken at the hospital for DNA profiling and comparison purposes. DNA was successfully extracted from the humerus bones and the buccal swabs taken, yielding 0.486 ng/μl, 0.294 ng/μl, and 0.621 ng/μl for human remains E9, E10 and E11 respectively as shown in table 1. Buccal swab samples of alleged relatives E1, E3, and E5 also yielded 12.872 ng/μl, 14.423 ng/μl, and 11.845 ng/μl, respectively. These values were enough to generate full DNA profiles for relationship testing.
The successful recovery of DNA from femur bone agrees with the results of other published research that suggested the viability of obtaining higher quantities of endogenous DNA for forensic cases by sampling the petrous bone, fingernails and femur bones for cases in which there is extensive degradation to hard tissues degraded skeletal remains [3,10,12]. DNA profiles generated from the humerus of human remains E9 and E10 did not match the DNA profile from an alleged relative from family A.
Full DNA profiles were successfully generated for the human remains, E11 and alleged surviving relatives of the deceased (E5, ie, child; and E3, ie, father) for relationship testing (Figure 2). Assuming a prior probability of 0.5, the combined paternity index was calculated to be 1.047 × 108, using published population data [13] and DNA View software [14]. This means the observed profile was 1.047 × 108 times more likely to occur under the scenario that the deceased is the true biological father of surviving relative (daughter), as opposed to the scenario that the deceased is an unrelated person of the Black African population [13] to the surviving relative (daughter). With a confidence probability of 99.99999904%, the conclusion is based on the calculated frequency of the DNA profile is very rare in unrelated individuals of the Black African Race.
The DNA analysis matched members of family B to human remains E11 and brought some closure to the case for that family.
Compliance with ethical standards
Funding: Authors received no financial support from any organization.
Research involving human participants and/oranimals:
Approval was obtained from the Ethics Committee of the Kwame Nkrumah University of Science and Technology, Kumasi.
Informed consent: Informed consent was obtained from the study participants.
Ethical approval: Ethical approval Committee of the Kwame Nkrumah University of Science and Technology, Kumasi.
The authors would like to thank the Management of Enchi Government Hospital for providing the samples, and the staff of the forensic science Laboratory for their support.
- Hebda L. DNA Isolation and Analysis from Skeletal Remains: Evaluating the Utility of Soil DNA Extraction Kits. Michigan State University. Forensic Science. 2013.
- Claridge J. Exhuming a Corpse for Forensic Analysis. 2016. http://www.exploreforensics.co.uk/
- Drobnic K. PCR Analysis of DNA from Skeletal Remains in Crime Investigation Cases. In Problems of Forensic Science, Special issue: Second European Academy of Forensic Science Meeting, Cracow. 2001; 46: 110-115.
- Truong L, Park HL, Chang SS, Ziogas A, Neuhausen SL, et al. Human Nail Clippings as a Source of DNA for Genetic Studies. Open J Epidemiol. 2015; 5: 41-50. PubMed: https://pubmed.ncbi.nlm.nih.gov/26180661/
- Hogervorst JGF, Godschalk RW, van den Brandt PA, Weijenberg MP, Verhage BA, et al. DNA from nails for genetic analyses in large-scale epidemiologic studies. Cancer Epidemiol Biomarkers Prev. 2014; 23: 2703-2712. PubMed: https://pubmed.ncbi.nlm.nih.gov/25472680/
- Primorac D, Andelinovic S, Defi ni-Gojanovic M, Drmic I, Rezic B, et al. Identification of war victims from mass graves in Croatia, Bosnia, and Herzegovina by the use of standard forensic methods and DNA typing. J Forensic Sci. 1996; 41: 891-894. https://bit.ly/3fdZyK
- ICRC. Missing people, DNA analysis and identification of human remains: A guide to best practice in armed conflicts and other situations of armed violence. 2nd edition 15-25. 2009. https://bit.ly/2KYtNqD
- Afrifah KA, Boateng AB, Akomeah SA, Motey EE, Abban EK, et al. Missing Persons Identification: Genetic profiling of highly charred human remains using sixteen STR loci markers. Forensic Sci Today. 2020; 6: 016-019. https://www.peertechzpublications.com/articles/FST-6-117.php
- Marjanović D, Metjahić HN, Čakar J, Džehverović M, Dogan S, et al. Identification of human remains from the Second World War mass graves uncovered in Bosnia and Herzegovina. Croat Med J. 2015; 56: 257-262. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4500967/
- Afrifah KA, Badu-Boateng A, Antwi-Akomeah S, Motey EE, Boampong E, et al. Forensic identification of missing persons using DNA from surviving relatives and femur bone retrieved from salty environment. J Forensic Sci Med. 2020; 6: 40-44. https://bit.ly/3dhZdo4
- Badu-Boateng A, Twumasi P, Salifu SP, Afrifah KA. A comparative study of different laboratory storage conditions for enhanced DNA analysis of crime scene soil-blood mixed sample. Forensic Sci Int. 2018; 292: 97-109. https://bit.ly/3fuQ11M
- Gaudio D, Fernandes DM, Schmidt R, Cheronet O, Mazzarelli D, et al. Genome-Wide DNA from degraded petrous bones and the assessment of sex and probable geographic origins of forensic cases. Sci Rep. 2019; 9: 8226. https://go.nature.com/2WGe5Gk
- Hares DR. Selection and Implementation of Expanded CODIS Core Loci in the United States. Forensic Sci Int Genet. 2015; 17: 33-34. Link: https://bit.ly/2WtBEln
- Brenner CH. Fundamental problem of forensic mathematics – the evidential value of a rare haplotype. Forensic Sci Int Genet. 2010; 4: 281-291. PubMed: https://pubmed.ncbi.nlm.nih.gov/20457055/