Abstract
Review Article
Biotechnology in Forensic Science: Advancements and Applications
Sunny Antil and Vandana Joon*
Published: 25 February, 2025 | Volume 9 - Issue 1 | Pages: 007-014
Background: Biotechnology is a multidisciplinary field based on the expertise of molecular biology, chemistry, biochemistry, chemical and biological engineering, and digital computing. Biotechnology plays an important role in modern forensic science, driving advances in analytical tools and techniques.
This review study provides a brief overview of applications, highlighting advances in forensic biotechnology and key technologies involved in the domains of genomics and DNA analysis, microbial forensics, forensic medicine, and forensic serology. The integration of forensic expertise with technology has increased the accuracy, sensitivity, and efficiency of forensic casework.
Conclusions: This interdisciplinary field extends beyond its usual association with biology to also include chemistry, fingerprint analysis, and toxicology, among others. Continued progress and innovation in this advanced field will further enhance investigative capabilities and facilitate the pursuit of justice.
Read Full Article HTML DOI: 10.29328/journal.jfsr.1001073 Cite this Article Read Full Article PDF
Keywords:
Body fluids identification; Forensics; FTIR; Next generation sequencing; NMR; Raman spectroscopy; Spectroscopic techniques
References
- Singh KR. A review paper on the techniques of biotechnology. Int J Emerg Technol Innov Res. 2018;5(9):233-238. Available from: http://www.jetir.org/papers/JETIRFH06040.pdf
- Verma AS, Agrahari S, Rastogi S, Singh A. Biotechnology in the realm of history. J Pharm Bioallied Sci. 2011;3(3):321-323. Available from: https://doi.org/10.4103/0975-7406.84430
- Agrawal M, Biswas S, Vliet K. Vacuum technology-assisted forensic biotechnology revealing latent fingerprints from fabrics. Vac Technol Coat. 2016;17:32-34. Available from: https://www.researchgate.net/publication/308314408_Vacuum_Technology_Assisted_Forensic_Biotechnology_Revealing_Latent_Finger_Prints_from_Fabrics
- Maras MH, Miranda M. Forensic science. In: Encyclopedia of Criminology and Criminal Justice. 2014;1-11. Available from: https://doi.org/10.1080/23779497.2017.1281088
- Shukla RK. Forensic biotechnology: Application of flow cytometry in legal medicine. Int J Forensic Sci. 2016. Available from: https://doi.org/10.23880/IJFSC-16000103
- Nizami BN, Kazmi SZH, Abid F, Babar MM, Noor A, Zaidi NUS, et al. Omics approaches in forensic biotechnology: looking for ancestry to offence. In: Barh D, Azevedo V, editors. Omics Technologies and Bio-Engineering. Academic Press. 2018:111-129. Available from: https://doi.org/10.1016/B978-0-12-804659-3.00006-3
- Bartelink EJ, Chesson LA. Recent applications of isotope analysis to forensic anthropology. Forensic Sci Res. 2019;4(1):29-44. Available from: https://doi.org/10.1080/20961790.2018.1549527
- Lewis M, Lainé K, Dawnay L, Lamont D, Scott K, Mariani S, et al. The forensic potential of environmental DNA (eDNA) in freshwater wildlife crime investigations: From research to application. Sci Justice. 2024;64(4):443-454. Available from: https://doi.org/10.1016/j.scijus.2024.06.003
- National Forensic Sciences University. Master's and specialization programs in forensic biotechnology. National Forensic Sciences University. [Accessed February 26, 2024]. Available from: https://www.nfsu.ac.in/Programs/programinfo/13?deptid=43
- Barkham T. BioRobot EZ1 workstation compares well with manual spin kits for extraction of viral RNA from sera and saves substantial staff time. J Clin Microbiol. 2006;44(4):1598. Available from: https://doi.org/10.1128/JCM.44.4.1598.2006
- National Institute of Justice. Automation of sexual assault DNA processing increases efficiency. National Institute of Justice. [Accessed February 26, 2024]. Available from: https://nij.ojp.gov/topics/articles/automation-sexual-assault-dna-processing-increases-efficiency
- QIAsymphony SP/AS. [Accessed February 26, 2024]. Available from: https://www.qiagen.com/us/products/automation/dna-rna-purification/qiasymphony-spas/
- Kaunitz JD. The discovery of PCR: ProCuRement of divine power. Dig Dis Sci. 2015;60(8):2230-2231. Available from: https://doi.org/10.1007/s10620-015-3747-0
- Pillay VV, Menezes RG, Krishnaprasad R, Pillay M, Lobo SW, Adhikari D, et al. Biotechnology in forensic science: the revolution continues. Nepal Med Coll J. 2007;9(1):57-62. Available from: https://pubmed.ncbi.nlm.nih.gov/17593681/
- Shrivastava P, Jain T, Kumawat RK. Direct PCR amplification from saliva sample using non-direct multiplex STR kits for forensic DNA typing. Sci Rep. 2021;11(1):7112. Available from: https://doi.org/10.1038/s41598-021-86633-0
- Imaizumi K, Noguchi K, Shiraishi T, Sekiguchi K, Senju H, Fujii K, et al. DNA typing of bone specimens—the potential use of the profiler test as a tool for bone identification. Leg Med (Tokyo). 2005;7(1):31-41. Available from: https://doi.org/10.1016/j.legalmed.2004.07.003
- Pereira R, Oliveira J, Sousa M. Bioinformatics and computational tools for next-generation sequencing analysis in clinical genetics. J Clin Med. 2020;9(1):132. Available from: https://doi.org/10.3390/jcm9010132
- Jager AC, Alvarez ML, Davis CP, Guzmán E, Han Y, Way L, et al. Developmental validation of the miSeq FGx forensic genomics system for targeted next-generation sequencing in forensic DNA casework and database laboratories. Forensic Sci Int Genet. 2017;28:52-70. Available from: https://doi.org/10.1016/j.fsigen.2017.01.011
- Martin P, de Simón LF, Luque G, Farfán MJ, Alonso A. Improving DNA data exchange: Validation studies on a single 6 dye STR kit with 24 loci. Forensic Sci Int Genet. 2014;13:68-78. Available from: https://doi.org/10.1016/j.fsigen.2014.07.002
- Oostdik K, Lenz K, Nye J, Schelling K, Yet D, Bruski S, et al. Developmental validation of the PowerPlex® Y23 System: A male-specific, Y-STR multiplex system. Forensic Sci Int Genet. 2014;12:77-88. Available from: https://doi.org/10.1016/j.fsigen.2014.04.013
- Whitaker JP, Clayton TM, Urquhart AJ, Millican ES, Downes TJ, Kimpton CP, et al. Short tandem repeat typing of bodies from a mass disaster: high success rate and characteristic amplification patterns in highly degraded samples. Biotechniques. 1995 Apr;18(4):670-7. Available from: https://pubmed.ncbi.nlm.nih.gov/7598902/
- Budowle B, Bieber FR, Eisenberg AJ. Forensic aspects of mass disasters: Strategic considerations for DNA-based human identification. Leg Med (Tokyo). 2005;7:230-243. Available from: https://doi.org/10.1016/j.legalmed.2005.01.001
- Hughes-Stamm SR, Ashton KJ, van Daal A. Assessment of DNA degradation and the genotyping success of highly degraded samples. Int J Legal Med. 2011;125:341-348. Available from: https://doi.org/10.1007/s00414-010-0455-3
- Alvarez-Cubero MJ, Saiz M, Martínez-García B, Sayalero SM, Entrala C, Lorente JA, et al. Next-generation sequencing: An application in forensic sciences? Ann Hum Biol. 2017;44(7):581-592. Available from: https://doi.org/10.1080/03014460.2017.1375155
- Haddrill PR. Developments in forensic DNA analysis. Emerg Top Life Sci. 2021;5(3):381-393. Available from: https://doi.org/10.1042/ETLS20200304
- Knijff P. From next-generation sequencing to now-generation sequencing in forensics. Forensic Sci Int Genet. 2019;38:175-180. Available from: https://doi.org/10.1016/j.fsigen.2018.10.017
- Davenport L, Devesse L, Syndercombe Court D, Ballard D. Forensic identity SNPs: Characterisation of flanking region variation using massively parallel sequencing. Forensic Sci Int Genet. 2023;64:102847. Available from: https://doi.org/10.1016/j.fsigen.2023.102847
- Novroski NMM, Cihlar JC. Evolution of single-nucleotide polymorphism use in forensic genetics. Forensic Sci Int Genet. 2022;4:6. Available from: https://doi.org/10.1002/wfs2.1459
- Pontes L, Sousa JC, Medeiros R. SNPs and STRs in forensic medicine: A strategy for kinship evaluation. Arch Med Sadowej Kryminol. 2017;67(3):226-240. Available from: https://doi.org/10.5114/amsik.2017.73194
- Yagasaki K, Mabuchi A, Higashino T, Wong JH, Nishida N, Fujimoto A, et al. Practical forensic use of kinship determination using high-density SNP profiling based on a microarray platform, focusing on low-quantity DNA. Forensic Sci Int Genet. 2022;61:102752. Available from: https://doi.org/10.1016/j.fsigen.2022.102752
- Zhang Q, Wang X, Cheng P, Yang S, Li W, Zhou Z, Wang S. Complex kinship analysis with a combination of STRs, SNPs, and indels. Forensic Sci Int Genet. 2022;61:102749. Available from: https://doi.org/10.1016/j.fsigen.2022.102749
- Dash HR, Arora M. CRISPR-CasB technology in forensic DNA analysis: Challenges and solutions. Appl Microbiol Biotechnol. 2022;106(12):4367-4374. Available from: https://doi.org/10.1007/s00253-022-12016-8
- Barash M, McNevin D, Fedorenko V, Giverts P. Machine learning applications in forensic DNA profiling: A critical review. Forensic Sci Int Genet. 2024;69:102994. Available from: https://doi.org/10.1016/j.fsigen.2023.102994
- Al-Hakim R, Putri E, Hidayah H, Pangestu A, Riani S. Current evidence on bioinformatics' role and digital forensics that contribute to forensic science: Upcoming threat. J Ris Rumpun Matematika Ilmu Pengetahuan Alam. 2022;1(1):25-32. Available from: https://doi.org/10.55606/jurrimipa.v1i1.157
- Ray PK. Bioinformatics as a modern tool in forensic science for data understanding & investigation in research. J Forensic Sci Res. 2022;6:83-87. Available from: https://doi.org/10.29328/journal.jfsr.1001040
- Wallace H, Jackson AR, Gruber J, Thibedeau AD. Forensic DNA databases: Ethical and legal standards: A global review. Egypt J Forensic Sci. 2014;4:57-63. Available from: https://doi.org/10.1016/j.ejfs.2014.04.002
- Behera C, Singh P, Shukla P, Bharti DR, Kaushik R, Sharma N, et al. Development of the first DNA database and identification portal for identification of unidentified bodies in India - UMID. Sci Justice. 2022;62(1):110-116. Available from: https://doi.org/10.1016/j.scijus.2021.12.002
- Chunkul S, Sathirapatya T, Dangklao P, Kawicha P, Tammachote R, Vongpaisarnsin K. Enhancing the forensic sexual assault investigations with LAMP-based male DNA detection. Forensic Sci Int Synergy. 2024;10:100567. Available from: https://doi.org/10.1016/j.fsisyn.2024.100567
- Guo X, Gu L, Luo Y, Wang S, Luo H, Song F. A bibliometric analysis of microbial forensics from 1984 to 2022: Progress and research trends. Front Microbiol. 2023;14:1186372. Available from: https://doi.org/10.3389/fmicb.2023.1186372
- Oliveira M, Amorim A. Microbial forensics: new breakthroughs and future prospects. Appl Microbiol Biotechnol. 2018;102(24):10377-10391. Available from: https://doi.org/10.1007/s00253-018-9414-6
- Zhang L, Chen F, Zeng Z, Xu M, Sun F, Yang L, et al. Advances in metagenomics and its application in environmental microorganisms. Front Microbiol. 2021;12:766364. Available from: https://doi.org/10.3389/fmicb.2021.766364
- Singh B, Publow A. Forensic body fluid identification using microbiome signature attribution. September 2020. 51 pages.
- Dobay A, Haas C, Fucile G, Downey N, Morrison HG, Kratzer A, et al. Microbiome-based body fluid identification of samples exposed to indoor conditions. Forensic Sci Int Genet. 2019;40:105-113. Available from: https://doi.org/10.1016/j.fsigen.2019.02.010
- Tozzo P, D'Angiolella G, Brun P, Castagliuolo I, Gino S, Caenazzo L. Skin microbiome analysis for forensic human identification: What do we know so far? Microorganisms. 2020;8(6):873. Available from: https://doi.org/10.3390/microorganisms8060873
- Cláudia-Ferreira A, Barbosa DJ, Saegeman V, Fernández-Rodríguez A, Dinis-Oliveira RJ, Freitas AR. On Behalf of the Escmid Study Group of Forensic and Post-Mortem Microbiology Esgfor. The Future Is Now: Unraveling the Expanding Potential of Human (Necro) Microbiome in Forensic Investigations. Microorganisms. 2023;11(10):2509. Available from: https://doi.org/10.3390/microorganisms11102509
- Nodari R, Arghittu M, Bailo P, Cattaneo C, Creti R, D'Aleo F, et al. ESCMID Study Group of Forensic and Post-Mortem Microbiology (ESGFOR) and the AMCLI Forensic Microbiology Study Group (GLAMIFO). Forensic Microbiology: When, Where and How. Microorganisms. 2024;12(5):988. Available from: https://doi.org/10.3390/microorganisms12050988
- Tejaswi KB, Hari Periya EA. Virtopsy (virtual autopsy): A new phase in forensic investigation. J Forensic Dent Sci. 2013;5(2):146-148. Available from: https://pubmed.ncbi.nlm.nih.gov/24255565/
- Turner S, Morrison S. Advances in forensic imaging technology for postmortem analysis. J Med Imaging Health Inform. 2016;6(5):1220-1228.
- Varnell RM, Stimac GK, Fligner CL. CT diagnosis of toxic brain injury in cyanide poisoning: Considerations for forensic medicine. Am J Neuroradiol. 1987;8(6):1063-1066. Available from: https://pubmed.ncbi.nlm.nih.gov/3120533/
- Ikeda T. Estimating age at death based on costal cartilage calcification. Tohoku J Exp Med. 2017;243:237-246. Available from: https://doi.org/10.1620/tjem.243.237
- Grabherr S, Baumann P, Minoiu C, Fahrni S, Mangin P. Post-mortem imaging in forensic investigations: Current utility, limitations, and ongoing developments. Res Rep Forensic Med Sci. 2016;6:25-37. Available from: https://doi.org/10.2147/RRFMS.S93974
- Gupta NS, Vandana, Rohatgi R, Gupta S. Title of the article. J Forensic Med Toxicol. 2022;39(2):97-104. Medico Legal Society.
- Boroumand M, Grassi VM, Castagnola F, De-Giorgio F, D’aloja E, Vetrugno G, et al. Estimation of postmortem interval using top-down HPLC–MS analysis of peptide fragments in vitreous humour: A pilot study. Int J Mass Spectrom. 2022;483:116952. Available from: https://doi.org/10.1016/j.ijms.2022.116952
- Zapico SC, Adserias-Garriga J. Postmortem interval estimation: New approaches by the analysis of human tissues and microbial communities’ changes. Forensic Sci. 2022;2:163-174. Available from: https://doi.org/10.3390/forensicsci2010013
- Choi KM, Zissler A, Kim E, Ehrenfellner B, Cho E, Lee SI, et al. Postmortem proteomics to discover biomarkers for forensic PMI estimation. Int J Legal Med. 2019;133(3):899-908. Available from: https://doi.org/10.1007/s00414-019-02011-6
- Locci E, Stocchero M, Gottardo R, De-Giorgio F, Demontis R, Nioi M, et al. Comparative use of aqueous humour 1H NMR metabolomics and potassium concentration for PMI estimation in an animal model. Int J Legal Med. 2021;135(3):845-852. Available from: https://doi.org/10.1007/s00414-020-02468-w
- Williams T, Soni S, White J, Can G, Javan GT. Evaluation of DNA degradation using flow cytometry: Promising tool for postmortem interval determination. Am J Forensic Med Pathol. 2015;36(2):104-110. Available from: https://doi.org/10.1097/PAF.0000000000000146
- Toma L, Vignali G, Maffioli E, Tambuzzi S, Giaccari R, Mattarozzi M, et al. Mass spectrometry-based proteomic strategy for ecchymotic skin examination in forensic pathology. Sci Rep. 2023;13(1):6116. Available from: https://doi.org/10.1038/s41598-023-32520-9
- Nagana-Gowda GA, Gowda YN, Raftery D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal Chem. 2015;87(1):706-715. Available from: https://doi.org/10.1021/ac503651e
- Carr S. Forensic science: The future of body fluid identification. Meas Control. 2009;42(10):310-313. Available from: https://doi.org/10.1177/002029400904201004
- Van-Steendam K, De Ceuleneer M, Dhaenens M, Van Hoofstat D, Deforce D. Mass spectrometry-based proteomics as a tool to identify biological matrices in forensic science. Int J Legal Med. 2013;127(2):287-298. Available from: https://doi.org/10.1007/s00414-012-0747-x
- Jackson S, Frey BS, Bates MN, Swiner DJ, Badu-Tawiah AK. Direct differentiation of whole blood for forensic serology analysis by thread spray mass spectrometry. Analyst. 2020;145(16):5615-5623. Available from: https://doi.org/10.1039/d0an00857e
- Muro CK, Doty KC, de Souza Fernandes L, Lednev IK. Forensic body fluid identification and differentiation by Raman spectroscopy. Forensic Chem. 2016;1:31-38. Available from: https://doi.org/10.1016/J.FORC.2016.06.003
- Hanson E, Ingold S, Haas C, Ballantyne J. Messenger RNA biomarker signatures for forensic body fluid identification revealed by targeted RNA sequencing. Forensic Sci Int Genet. 2018;34:206-221. Available from: https://doi.org/10.1016/j.fsigen.2018.02.020
- Tighe PJ, Ryder RR, Todd I, Fairclough LC. ELISA in the multiplex era: Potentials and pitfalls. Proteomics Clin Appl. 2015;9(3-4):406-422. Available from: https://doi.org/10.1002/prca.201400130
- Bazyar H. On the Application of Microfluidic-Based Technologies in Forensics: A Review. Sensors (Basel, Switzerland). 2023;23(13):5856. Available from: https://doi.org/10.3390/s23135856
- Medina-Paz F, Kuba B, Kryvorutsky E, Roca G, Zapico SC. Assessment of blood and semen detection and DNA collection from swabs up to three months after deposition on five different cloth materials. Int J Mol Sci. 2024;25(6):3522. Available from: https://doi.org/10.3390/ijms25063522
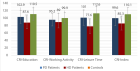
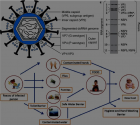
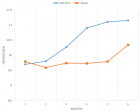
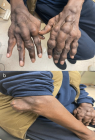
Figures:

Figure 1
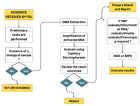
Figure 2

Figure 3
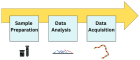
Figure 4
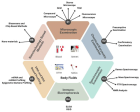
Figure 5
Similar Articles
-
Awareness level on the role of forensic DNA database in criminal investigation in Nigeria: A case study of Benin cityNwawuba Stanley Udogadi*,Akpata Chinyere Blessing Nkiruka . Awareness level on the role of forensic DNA database in criminal investigation in Nigeria: A case study of Benin city. . 2020 doi: 10.29328/journal.jfsr.1001019; 4: 007-014
-
Awareness level on the relevance of forensics in criminal investigation in NigeriaOmorogbe Owen Stephen,Orhue Osazee Kelvin,Ehikhamenor Edeaghe,Nwawuba Stanley Udogadi*. Awareness level on the relevance of forensics in criminal investigation in Nigeria. . 2021 doi: 10.29328/journal.jfsr.1001028; 5: 053-057
-
Comparative analysis of mobile forensic proprietary tools: an application in forensic investigationParth Chauhan*,Tamanna Jaitly,Animesh Kumar Agrawal. Comparative analysis of mobile forensic proprietary tools: an application in forensic investigation. . 2022 doi: 10.29328/journal.jfsr.1001039; 6: 077-082
-
Forensic Analysis of WhatsApp: A Review of Techniques, Challenges, and Future DirectionsNishchal Soni*. Forensic Analysis of WhatsApp: A Review of Techniques, Challenges, and Future Directions. . 2024 doi: 10.29328/journal.jfsr.1001059; 8: 019-024
-
Analysis and Comparison of Social Media Applications Using Forensic Software on Mobile DevicesHüseyin Çakır*, Merve Hatice Karataş . Analysis and Comparison of Social Media Applications Using Forensic Software on Mobile Devices. . 2024 doi: 10.29328/journal.jfsr.1001065; 8: 058-063
-
Biotechnology in Forensic Science: Advancements and ApplicationsSunny Antil,Vandana Joon*. Biotechnology in Forensic Science: Advancements and Applications. . 2025 doi: 10.29328/journal.jfsr.1001073; 9: 007-014
-
Digital Forensics and Media Offences – Investigate Synergy in the Cyber AgeGauri Goyal*. Digital Forensics and Media Offences – Investigate Synergy in the Cyber Age. . 2025 doi: 10.29328/journal.jfsr.1001074; 9: 015-020
Recently Viewed
-
Clinical and Histopathological Mismatch: A Case Report of Acral FibromyxomaMonica Mishra*,Kailas Mulsange,Gunvanti Rathod,Deepthi Konda. Clinical and Histopathological Mismatch: A Case Report of Acral Fibromyxoma. Arch Pathol Clin Res. 2025: doi: 10.29328/journal.apcr.1001045; 9: 005-007
-
Unconventional powder method is a useful technique to determine the latent fingerprint impressionsHarshita Niranjan,Shweta Rai,Kapil Raikwar,Chanchal Kamle,Rakesh Mia*. Unconventional powder method is a useful technique to determine the latent fingerprint impressions. J Forensic Sci Res. 2022: doi: 10.29328/journal.jfsr.1001035; 6: 045-048
-
Doppler Evaluation of Renal Vessels in Pediatric Patients with Relapse and Remission in Different Categories of Nephrotic SyndromeAmit Nandan Dhar Dwivedi*, Srishti Sharma, OP Mishra, Girish Singh. Doppler Evaluation of Renal Vessels in Pediatric Patients with Relapse and Remission in Different Categories of Nephrotic Syndrome. J Clini Nephrol. 2023: doi: 10.29328/journal.jcn.1001112; 7: 067-072
-
Atlantoaxial subluxation in the pediatric patient: Case series and literature reviewCatherine A Mazzola*,Catherine Christie,Isabel A Snee,Hamail Iqbal. Atlantoaxial subluxation in the pediatric patient: Case series and literature review. J Neurosci Neurol Disord. 2020: doi: 10.29328/journal.jnnd.1001037; 4: 069-074
-
Intelligent Design of Ecological Furniture in Risk Areas based on Artificial SimulationTorres del Salto Rommy Adelfa*, Bryan Alfonso Colorado Pástor*. Intelligent Design of Ecological Furniture in Risk Areas based on Artificial Simulation. Arch Surg Clin Res. 2024: doi: 10.29328/journal.ascr.1001083; 8: 062-068
Most Viewed
-
Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth EnhancersH Pérez-Aguilar*, M Lacruz-Asaro, F Arán-Ais. Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth Enhancers. J Plant Sci Phytopathol. 2023 doi: 10.29328/journal.jpsp.1001104; 7: 042-047
-
Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case PresentationJulian A Purrinos*, Ramzi Younis. Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case Presentation. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001099; 8: 075-077
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Pediatric Dysgerminoma: Unveiling a Rare Ovarian TumorFaten Limaiem*, Khalil Saffar, Ahmed Halouani. Pediatric Dysgerminoma: Unveiling a Rare Ovarian Tumor. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001087; 8: 010-013
-
Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative reviewKhashayar Maroufi*. Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative review. J Sports Med Ther. 2021 doi: 10.29328/journal.jsmt.1001051; 6: 001-007

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."