Abstract
Research Article
Rapid and sensitive identification of cow and buffalo species and gender in tissue/meat samples impounded from different spots in Delhi NCR India by Real Time PCR
Naresh Kumar*, Rahul Jain, Seerat, Amit Chauhan and Deepa Verma
Published: 12 January, 2022 | Volume 6 - Issue 1 | Pages: 012-016
The objective of this study was to obtain a fast, accurate and reliable method of species identification of unknown biological samples for forensic applications, especially in illegal trade of animals as well as meat fraud. Meat fraud and adulteration not only affects the market but also increases the risk of religious and ethnic conflicts around the world [1]. In this study, species-specific and gender differentiating Real time PCR technique was employed to analyse 15 meat samples collected from a suspected site. Out of 15 samples collected from suspected site, 54% and 13% samples were of Cow and buffalo origin respectively. All 54% cow samples were of male while one each of buffalo were of male and female origin. Two samples were inconclusive. These findings indicated that species and gender-specific PCR is very sensitive and can be used for forensic species identification and the detection of meat fraud and adulteration.
Read Full Article HTML DOI: 10.29328/journal.jfsr.1001031 Cite this Article Read Full Article PDF
Keywords:
Real time PCR; Adulteration; Species identification; mtDNA; DNA testing; Meat labeling
References
- O’Mahony PJ. Finding horse meat in beef products–a global problem. 2013; 106: 595–597.
- Malnekoff E. Cattle Smuggling from India to Bangladesh. Honors Theses. 2013; scholarworks.wmich.edu/honors-theses/
- Kumar D, Singh SP, Karabasanavar NS, Singh R, Umapathi V.Authentication of beef, carabeef, chevon, mutton and pork by a PCR-RFLP assay of mitochondrial cytb gene. J Food Sci Technol. 2014; 51: 3458–3463. PubMed: https://pubmed.ncbi.nlm.nih.gov/26396346/
- Mane BG, Mendiratta SK, Tiwari AK, Bhilegaokar KN. Detection of Adulteration of Meat and Meat Products with Buffalo Meat Employing Polymerase Chain Reaction Assay. Food Anal Methods. 2012; 5: 296–300.
- Haider N, Nabulsi I, Al-Safadi B. Identifification of meat species by PCR-RFLP of the mitochondrial COI gene. Meat Sci. 2012; 90: 490–493. PubMed: https://pubmed.ncbi.nlm.nih.gov/21996288/
- Lou XP, Zhang W, Zheng J, Xu H, Zhao F. Comparative study on morphology of human, swine, sheep and cattle muscle tissues and its forensic significance.Fa Yi Xue Za Zhi. 2016; 32: 250–253. PubMed: https://pubmed.ncbi.nlm.nih.gov/29188664/
- Haunshi S, Basumatary R, Girish PS, Doley S, Bardoloi RK, et al. Identifification of chicken, duck, pigeon and pig meat by species-specifific markers of mitochondrial origin. Meat Sci. 2009; 83: 454–459. PubMed: https://pubmed.ncbi.nlm.nih.gov/20416682/
- Ishida N, Sakurada M, Kusunoki H, Ueno Y. Development of a simultaneous identification method for 13 animal species using two multiplex real-time PCR assays and melting curve analysis. Leg Med. 2018; 30: 64-71. PubMed: https://pubmed.ncbi.nlm.nih.gov/29197713/
- Rastogi G, Dharne MS, Walujkar S, Kumar A, Patole MS, et al. Species identifification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 2007; 76: 666–674. PubMed: https://pubmed.ncbi.nlm.nih.gov/22061243/
- Lockley AK, Bardsley RG. DNA-based methods for food authentication. Trends in Food Science & Technol. 2000; 11: 67-77.
- Hsieh HM, Chiang HL, Tsai LC, Lai SY, Huang NE, et al. Cytochrome b gene for species identification of the conservation animals. Forensic Sci Int. 2001; 122: 7-18. PubMed: https://pubmed.ncbi.nlm.nih.gov/11587860/
- Ennis S, Gallagher TF. A PCR-based sex-determination assay in cattle based on the bovine amelogenin locus. Animal Genetics. 1994; 25: 425-427. PubMed: https://pubmed.ncbi.nlm.nih.gov/7695123/
- Hsieh YH, Sheu SC, Bridgman RC. Development of a monoclonal antibody specific to cooked mammalian meats. J Food Prot. 1998; 61: 476- 481. PubMed: https://pubmed.ncbi.nlm.nih.gov/9709213/
- Skarpeid HJ, Kvaal K, Hildrum KI. Identification of animal species in ground meat mixtures by multivariate analysis of isoelectricfocusing protein profiles. Electrophoresis. 1998; 19: 3103-3109. PubMed: https://pubmed.ncbi.nlm.nih.gov/9932802/
- Ebbehøj KF, Thomsen PD. Species differentiation of heated products by DNA hybridization. Meat Sci. 1991; 30: 221-234. PubMed: https://pubmed.ncbi.nlm.nih.gov/22061971/
- Janssen FW, Hägele GH, Buntjer JB, Lenstra JA. Species identification in meat by using PCR-generated satellite probes. J Industr Microbiol Biotechnol. 1998; 21: 115-120.
- Wolf C, Rentsch J, Hubner P. PCR-RFLP analysis of mitochondrial DNA: a reliable method for species identification. J Agric Food Chem. 1999; 47: 1350–1355. PubMed: https://pubmed.ncbi.nlm.nih.gov/10563979/
- Girish PS, Anjaneyulu ASR, Viswas KN, Shivakumar BM, Anand M, et al. Meat species identifification by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of mitochondrial 12S rRNA gene. Meat Sci. 2005l; 70: 107–112. PubMed: https://pubmed.ncbi.nlm.nih.gov/22063286/
- Maede D. A strategy for molecular species detection in meat and meat products by PCR-RFLP and DNA sequencing using mitochondrial and chromosomal genetic sequences. Eur Food Res Technol. 2006; 224: 209–217.
- Sebastio P, Zanelli P, Neri TM. Identification of anchovy (Engraulisencrasicholus L.) and gilt sardine (Sardinella aurita) by polymerase chain reaction, sequence of their mitochondrial cytochrome b gene, and restriction analysis of polymerase chain reaction products in semi preserves. J Agric Food Chem. 2001; 49: 1194-1199. PubMed: https://pubmed.ncbi.nlm.nih.gov/11312834/
- Asensio L, González I, Fernández A, Céspedes A, Rodríguez MA, et al. Identification of Nile perch (Lates niloticus), grouper (Epinephelusguaza), and wreck fish (Polyprion americanus) by polymerase chain reaction-restriction fragment length polymorphism of a 12S rRNA gene fragment. J Food Protection. 2000; 63: 1248-1252.
- Meyer R, Hoefelein C, Luethy J, Candrian U. Polymerase chain reaction-restriction fragment length polymorphism analysis: a simple method for species identification in food. Detection of soya in processed meat products. Z Lebensm Unters Forch. 1995;203: 339–344.
- Jonker K, Tilburg J, Hagele G, de Boer E. Species identification in meat products using real-time PCR. Food Additives and Contaminants. 2008; 25: 527-533.
- Lopparelli RM, Cardazzo B, Balzan S, Giaccone V, Novelli E. Real-time TaqMan polymerase chain reaction detection and quantification of cow DNA in pure water buffalo mozzarella cheese: method validation and its application on commercial samples. J Agric Food Chem. 2007; 55: 3429-3434. PubMed: https://pubmed.ncbi.nlm.nih.gov/17419643/
- Aasen E, Medrano J. Amplification of the Zfy and Zfx Genes for Sex Identification in Humans, Cattle, Sheep and Goats. Nat Biotechnol. 1990;8: 1279–1281. PubMed: https://pubmed.ncbi.nlm.nih.gov/1369448/
- Shi L, Yue W, Ren Y, Lei F, Zhao J. Sex determination in goat by amplification of the HMG box using duplex PCR. Animal Reproduction Science. 2008; 105: 398–403. PubMed: https://pubmed.ncbi.nlm.nih.gov/18096334/
- 2002.
- Ong SB, Zuraini MI, Jurin WG, Cheah YK, Tunung R, et al. Meat molecular detection: sensitivity of polymerase chain reaction-restriction fragment length polymorphism in species differentiation of meat from animal origin. ASEAN Food J. 2007; 14: 51–59.
- Luo J, Wang J, Bu D, Li D, Wang L, et al. Development and application of a PCR approach for detection of beef, sheep, pig, and chicken derived materials in feedstuff. Agr Sci China. 2008; 7: 1260–1266.
- Kumar A, Kumar RR, Sharma B, Gokulakrishnan P, Mendiratta SH, et al.Identification of Species Origin of Meat and Meat Products on the DNA Basis: A Review, Critical Reviews in Food Science and Nutrition. 2015; 55: 1340-1351. PubMed: https://pubmed.ncbi.nlm.nih.gov/24915324/
- Bellisa C, Ashtona KJ, Freneyb L, Blairb B, Griffifiths LR. A molecular genetic approach for forensic animal species identification. Forensic Sci Int. 2003; 134: 99–108. PubMed: https://pubmed.ncbi.nlm.nih.gov/12850402/
Figures:
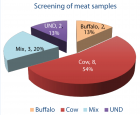
Figure 1

Figure 2
Similar Articles
-
Rapid and sensitive identification of cow and buffalo species and gender in tissue/meat samples impounded from different spots in Delhi NCR India by Real Time PCRNaresh Kumar*,Rahul Jain,Seerat,Amit Chauhan,Deepa Verma. Rapid and sensitive identification of cow and buffalo species and gender in tissue/meat samples impounded from different spots in Delhi NCR India by Real Time PCR. . 2022 doi: 10.29328/journal.jfsr.1001031; 6: 012-016
-
Bioinformatics as a modern tool in forensic science for data understanding & investigation in researchPranav Kumar Ray*. Bioinformatics as a modern tool in forensic science for data understanding & investigation in research. . 2022 doi: 10.29328/journal.jfsr.1001040; 6: 083-087
-
Forensic analysis of raw meat adulteration using mtDNAMarium Zehra*,Rukhsana Parveen,Muhammad Irfan,Mahrukh Nasir,Sidra Bashir. Forensic analysis of raw meat adulteration using mtDNA. . 2022 doi: 10.29328/journal.jfsr.1001041; 6: 088-092
Recently Viewed
-
Metastatic Brain Melanoma: A Rare Case with Review of LiteratureNeha Singh,Gaurav Raj,Akshay Kumar,Deepak Kumar Singh,Shivansh Dixit,Kaustubh Gupta*. Metastatic Brain Melanoma: A Rare Case with Review of Literature. J Radiol Oncol. 2025: doi: 10.29328/journal.jro.1001080; 9: 050-053
-
Validation of Prognostic Scores for Attempted Vaginal Delivery in Scar UterusMouiman Soukaina*,Mourran Oumaima,Etber Amina,Zeraidi Najia,Slaoui Aziz,Baydada Aziz. Validation of Prognostic Scores for Attempted Vaginal Delivery in Scar Uterus. Clin J Obstet Gynecol. 2025: doi: 10.29328/journal.cjog.1001185; 8: 023-029
-
Scientific Analysis of Eucharistic Miracles: Importance of a Standardization in EvaluationKelly Kearse*,Frank Ligaj. Scientific Analysis of Eucharistic Miracles: Importance of a Standardization in Evaluation. J Forensic Sci Res. 2024: doi: 10.29328/journal.jfsr.1001068; 8: 078-088
-
A study of coagulation profile in patients with cancer in a tertiary care hospitalGaurav Khichariya,Manjula K*,Subhashish Das,Kalyani R. A study of coagulation profile in patients with cancer in a tertiary care hospital. J Hematol Clin Res. 2021: doi: 10.29328/journal.jhcr.1001015; 5: 001-003
-
Additional Gold Recovery from Tailing Waste By Ion Exchange ResinsAshrapov UT*, Malikov Sh R, Erdanov MN, Mirzaev BB. Additional Gold Recovery from Tailing Waste By Ion Exchange Resins. Int J Phys Res Appl. 2024: doi: 10.29328/journal.ijpra.1001098; 7: 132-138
Most Viewed
-
Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth EnhancersH Pérez-Aguilar*, M Lacruz-Asaro, F Arán-Ais. Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth Enhancers. J Plant Sci Phytopathol. 2023 doi: 10.29328/journal.jpsp.1001104; 7: 042-047
-
Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case PresentationJulian A Purrinos*, Ramzi Younis. Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case Presentation. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001099; 8: 075-077
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Pediatric Dysgerminoma: Unveiling a Rare Ovarian TumorFaten Limaiem*, Khalil Saffar, Ahmed Halouani. Pediatric Dysgerminoma: Unveiling a Rare Ovarian Tumor. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001087; 8: 010-013
-
Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative reviewKhashayar Maroufi*. Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative review. J Sports Med Ther. 2021 doi: 10.29328/journal.jsmt.1001051; 6: 001-007

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."